On March 15, 2019, Shi Yigong, a research group of high-precision and innovative science in structural biology at Tsinghua University, published a study on the mechanism and structure of splices. In the journal Cell, the yeast splicing body with catalytic activation was published. "Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching", revealing the transient state of the first step of the splice splicing reaction - catalytically activated Spliceosome, defined as the "B* complex") high-resolution three-dimensional structure of four different conformations, which is the last unresolved basic state in the current RNA splicing cycle. So far, Shi Yigong has been the world's first and only team to successfully capture and analyze the results of a series of high-resolution three-dimensional structures of all fully assembled splices in the RNA splicing process. The four splice structures with the same conformation but different conformations reported in this paper have an overall resolution of 2.9 angstroms to 3.8 angstroms and a resolution of up to 2.7 angstroms in the core region. This is the highest resolution splice structure currently reported. It reveals the dynamic changes in the first step of the splicing reaction, and demonstrates the important role of splicing factors in the splicing reaction. The first time from the structural information, it is important to answer the specificity of the splices for different pre-mRNA substrate recognition. Scientific question.
In 1977, scientists first discovered that mRNA from adenovirus and its corresponding DNA transcriptional template could not form a continuous hybrid duplex, but extended a circular DNA single strand at different positions of the hybrid duplex. This major finding suggests that genetic information is transmitted from DNA to mRNA not only through transcription, but also requires pre-mRNA splicing to further complete the removal of "invalid" genetic information and the splicing of effective genetic information. The "invalid" genetic information does not have a translation function and is called an intron. The effective genetic information that can be translated by ribosomes is called an exon. The intron is removed and the exon is connected. This is RNA. Splicing. RNA splicing is ubiquitous in eukaryotes. As the species evolves, the number of genes containing introns increases, and the frequency of RNA splicing increases accordingly, making it possible for one gene to encode multiple proteins, greatly enriching proteins. The diversity of the group is also one of the important reasons for the diversity of eukaryotes.
RNA splicing is one of the important links in the regulation of eukaryotic gene expression, and its chemical nature is a two-step transesterification reaction. This seemingly simple chemical reaction is difficult to self-generate in cells, and the chemical reaction responsible for this is a large and highly dynamic molecular machine in the nucleus, the spliceosome. From the first discovery of RNA splicing in 1977 to the beginning of this century, scientists have established the process of assembly, activation and depolymerization of splices by immunoprecipitation, gene knockout, cross-linking mass spectrometry, and establishment of in vitro splicing reaction systems. And complex RNA splicing regulatory networks such as protein-protein, protein-nucleic acid interactions, and mutual regulation. During the splicing reaction, multiple protein-nucleic acid complexes and splicing factors are combined, rearranged and depolymerized in a highly precise sequence, which in turn forms a pre-assembled complex U4/U6.U5 Tri-snRNP (U4/U6.U5 III) The small nuclear ribonucleoprotein complex) and the fully assembled splice bodies at least 8 states pre-B, B, Bact, B*, C, C*, P, and ILS complexes (Fig. 1).

Figure 1 Schematic diagram of RNA splicing (Source: Yan, C., Wan, R., & Shi, Y. (2019). Cold Spring Harbor perspectives in biology, 11(1), a032409.)
Due to the dynamics and complexity of the splice height, the high resolution three-dimensional structure of the splice body in different states is recognized as a world problem. Under such a huge challenge, Professor Shi Yigong led the research team to overcome difficulties. After 7 years of hard work, the high-resolution structure of the fission yeast splice 3.6 angstrom was first reported in 2015, and the splicing was first demonstrated. The near-atomic resolution structure of the bulk catalytic center. This major research has revolutionized the study of RNA splicing mechanisms. Since the publication of the first splice structure in 2015, Shi Yigong has successively analyzed the high-resolution structures of S. cerevisiae splice complexes in eight different states, namely 3.8 angstroms of pre-assembled composite U4/U6. U5 Tri-snRNP, pre-catalyzed splice precursor pre-B complex of 3.3 angstroms to 4.6 angstroms, pre-catalyzed splicing body B complex of 3.9 angstroms, 3.5 angstrom activation state splicing body Bact complex, first step catalytic reaction of 3.4 angstroms The second splicing C complex, 4.0 angstrom second catalytically activated splice C* complex, 3.6 angstroms completed the two-step transesterification of the splice P complex, and the 3.5 angstrom intron lasso splicing ILS complex structure. These resolved splices cover the entire RNA splicing cycle, revealing the working mechanism of the splice-catalyzed two-step reaction of RNA splicing from the molecular level, and providing a structural basis for understanding the assembly, activation and depolymerization of splices. However, the last unresolved fully assembled splice B* complex is critical to understanding the occurrence of the first splicing reaction. Due to its high dynamic and transient nature, how to capture B* complex has become a major problem in the field. It is also one of the problems solved by different research groups in the world. It is extremely urgent to capture and analyze its structure.
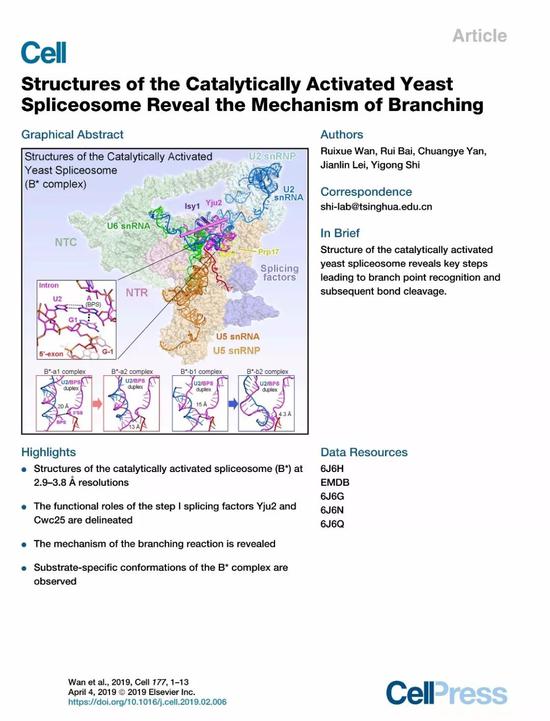
Despite the accumulated experience in the study of splice structure, Shi Yigong's research team is struggling and difficult to grasp how to capture a stable B* complex. Since B* complex and C complex are the two states in which the first splicing reaction occurs one after the other, the core difference in the identification of the domain is whether the phosphodiester bond between the 5' splice site and the branch point is formed. . Therefore, without affecting the normal assembly and activation of the splice body, how to prevent the first step reaction from occurring, and keep the 5' splice site and the branch point into the active reaction center of RNA splicing, and form the active site before the reaction It has become the biggest difficulty in this study. Shi Yigong's research team tried to directly block endogenous sources in vivo and block the splicing reaction in vitro. Finally, stable B* complex samples could not be obtained. In the newly published "Cell" article, Shi Yigong's research team combined in vivo blocking with the establishment of a splicing reaction in vitro, and repeatedly explored the purification program, and finally optimized a set of stable, good B. * complex sample. In order to explore the recognition specificity of the splices for different pre-mRNA substrates, Shi Yigong selected two pre-mRNAs commonly used in the field as substrates for splice-binding, and then reconstructed by single-particle cryo-electron microscopy. Four different conformations B* complex with a total resolution of 2.9 angstroms to 3.8 angstroms were constructed and an atomic model was constructed containing 4 RNAs and 35 proteins (Fig. 2).

Figure 2 Three-dimensional structure of four conformations of S. cerevisiae catalyzed activation of splices
The B* complex structure of four different conformations resolved in this paper demonstrates for the first time how the branch points in the first step of RNA splicing are gradually “pushed†into the active reaction center by the splicing body. The first step is the splicing factor. Yju2 plays a pivotal role in stabilizing branch points, although the distance between the 5' splice site and the branch point is very close (4.3 angstroms), but not enough to form a phosphodiester bond. Thus, this structure also clearly reveals the important role of the key protein factor Cwc25 in stimulating the first splicing reaction (Fig. 3). It is worth mentioning that in this structure, the mechanism that the branch point A in the pre-mRNA is recognized by the 5' splice site GU through the classical base complementary pairing and the base stacking force is observed for the first time. The importance of this site in the highly conserved nature of eukaryotes is further demonstrated. Another highlight of this paper is the first disclosure of the specificity of splices for different pre-mRNA substrate recognition, providing the most direct structural information for the differences in the splicing efficiency of different pre-mRNAs in vivo.
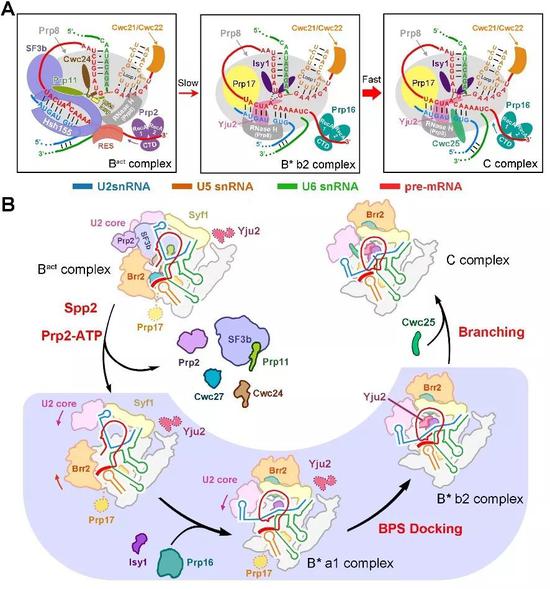
Figure 3 Model of Saccharomyces cerevisiae splice-mediated branching response
Up to now, Shi Yigong has analyzed a high-resolution three-dimensional structure of splices in 10 different states in yeast (Fig. 4). The results are all published in the top international journals Science and Cell (Science 7). Cell 3), from assembly to activation, from the occurrence of a two-step transesterification reaction to the depolymerization of the splice, the 10 states of the splices completely cover the splicing pathway, and the first splicing-mediated RNA splicing process is complete. The concatenation provides the clearest and most comprehensive structural information for understanding the molecular mechanisms of RNA splicing.
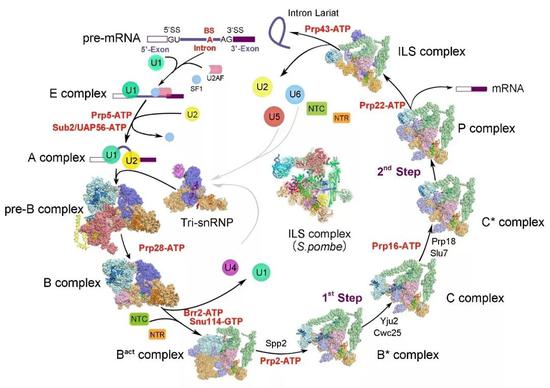
Figure 4 Summary of yeast splice structure analyzed by the research group (Source: https://ygshi.org/research)
Ten years of a sword! Shi Yigong's laboratory on the study of splice structure reveals the molecular mechanism of RNA splicing. Professor Shi Yigong, the director of the Center for Structural Biology, Tsinghua University, is the author of this article; Tsinghua University School of Medicine, postdoctoral, highly sophisticated innovation center, excellence scholar Ruirui, Ph.D., a fourth-year doctoral student at the School of Life Sciences, is the co-first author of the article. Yan Chuang, a post-doctoral researcher at the Tsinghua University School of Life, and a brilliant scholar at the Center for Excellence in Innovation, helped to build the structural model; the director of the cryo-electron microscope platform at Tsinghua University Dr. Lei Jianlin provided assistance in the collection of cryo-electron microscopy data. The electron microscope data was collected from the cryo-electron microscope platform of Tsinghua University. The calculation work was supported by Tsinghua University High Performance Computing Platform and National Protein Facility Experimental Technology Center (Beijing). This work was supported by the Beijing Center for Structural Biology and Advanced Innovation (Tsinghua) and the National Natural Science Foundation.
Original link https://(19)30155-2
Related paper links http://science.sciencemag.org/content/early/2016/01/06/science.aad6466
Http://science.sciencemag.org/content/early/2015/08/19/science.aac8159
Http://science.sciencemag.org/content/early/2015/08/19/science.aac7629
Http://science.sciencemag.org/content/early/2016/07/20/science.aag0291
Http://science.sciencemag.org/content/early/2016/07/20/science.aag2235
Http://science.sciencemag.org/content/early/2016/12/14/science.aak9979.full
http://(17)30954-6
http://(17)31264-3
Http://science.sciencemag.org/content/early/2018/05/23/science.aau0325
Source: Highly sophisticated innovation center for structural biology
Prostate Treatment Machine
Prostate Treatment Machine is the most advanced health care device of the rostate gland in the world today. This product starts the new technologic era in the health care of the prostate gland. It combines far infrared ray, electronic pulse, magnet therapy and thermotherapy to improve the whole environment of the pelvic cavity in order to increase blood circulation of overall pelvic cavity.
It allows increase blood flow into the weakened tissue for enhance ecovery and pain relief. The special care to the most important accupoint [Huiyin" of prostate gland and pelvic cavity plays a very important role in the device, this helps to improve the total conditions of the procreation system. Meanwhile, this device also has obvious effects upon female pelvic cavity. Also it can help to prevent the disease of rectum.


Health Care Device,Prostate Treatment Machine,Prostate Gland Therapy,Prostate Gland Therapy Device
Shenzhen Guangyang Zhongkang Technology Co., Ltd. , https://www.syztreatment.com